Abstract
Background: Mixed phenotype acute leukemia (MPAL) is a subtype of leukemia with co-expression of lymphoid and myeloid markers. Genetic alterations are critical for the understanding of pathogenesis, diagnosis, risk stratification and treatment of hematological malignancies. Hitherto, in addition to t(9;22)(q34;q22)/BCR::ABL1 (Ph+) and KMT2A-rearrangements (KMT2A-r), BCL11B and ZNF384 rearrangements have been incorporated into the diagnosis and classification of MPAL. The genetic features in the remaining MPAL merit further investigation. This study aims to apply an integrated genomic and transcriptomic approach to explore the molecular characteristics of adult patients with MPAL.
Methods: 176 consecutive cases diagnosed with MPAL at the First Affiliated Hospital of Soochow University between January 2006 and November 2019 were included. Patients’ median age was 48 years old (range:16-87). The diagnoses were made according to the WHO 2016 criteria, 86 (48.9%) cases fulfilled T/My MPAL, NOS, 42 (23.9%) cases fulfilled Ph+ MPAL, 36 (20.5%) cases fulfilled B/My MPAL, NOS, 4 (2.3%) cases fulfilled MPAL with KMT2A-r, and 8 cases (4.5%) fulfilled MPAL, NOS, rare types. We performed targeted sequencing in 104 patients, RNA sequencing in 89 patients and whole genome sequencing in 4 MPAL patients. Gene expression profile data were compared to data from 133 AML, 53 B-ALL and 41 T-ALL cases. 120 of the patients who were treated in our hospital were applied for analysis of treatment response and survival.
Results: In total, 74 genes were found mutated with variant allele frequency more than 2% in 94 of 104 (90.4%) MPAL patients, with a total of 239 SNVs and 135 indels. Distinct mutational patterns were observed among different MPAL subtypes. Mutations of CEBPA (41.5% vs. 2.0%, P< 0.0001), DNMT3A (34.0% vs. 0%, P< 0.0001) and NOTCH1 (26.4% vs. 9.8%, P= 0.028) were mostly associated with T/My MPAL compared with in other MPALs. RUNX1 mutation was more common in Ph+ MPAL (51.9% vs. 7.8%, P< 0.0001). NOTCH1 mutation associated with B/T MPAL (60.0% vs. 16.2%, P= 0.041).
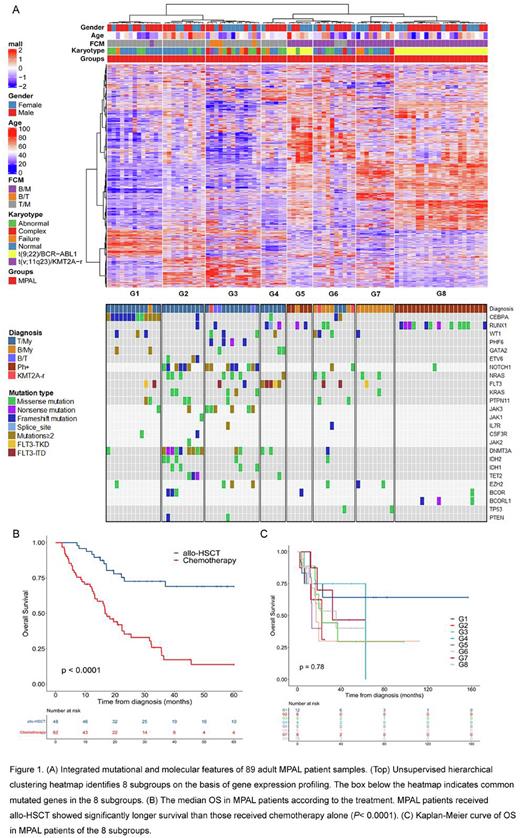
In comparison to other subtypes of leukemia, 33 of the 89 (37.1%) MPAL cases clustered with B-ALL, 30 (33.7%) clustered with T-ALL, and 26 (29.2%) clustered with AML. By integrating targeted sequencing and RNA sequencing data in 89 of the MPAL patients, we defined 8 molecular subgroups (G1-G8) with distinct mutational and gene expression characteristics (Figure 1A). G1 was associated with CEBPA mutations, G2 and G3 with NOTCH1 mutations, G4 with BCL11B rearrangement and FLT3 mutations, G5 and G8 with BCR::ABL1 fusion, G6 with KMT2A rearrangement/KMT2A rearrangement-like features, and G7 with ZNF384-rearrangement/ZNF384-rearrangement-like features.
With a median follow-up of 49.3 months, the estimated 5-year overall survival (OS) and event-free survival (EFS) in the whole cohort were 37.1 ± 5.1% and 27.9 ± 4.5%, respectively. There was no statistically significant difference in 5-year OS and EFS among patients with T/My, Ph+ and B/My MPAL. Of note, patients receiving allogeneic hematopoietic stem cell transplantation (allo-HSCT) had longer 5-year OS (69.1 ± 7.4% vs. 13.8 ± 5.6%, P< 0.0001, Figure 1B) and EFS (56.2 ± 7.5% vs. 7.4 ± 3.9%, P< 0.0001) than those receiving chemotherapy alone. Moreover, patients receiving allo-HSCT at first complete remission (CR1) had longer 5-year OS than those transplanted at non-CR1 (68.4 ± 11.3% vs. 52.7 ± 14.1%, P= 0.042). There was no significantly difference in the 5-year OS and EFS among the 8 subgroups as a whole (Figure 1C), which might due to the limited number of patients in each subgroup.
Conclusions: In this study, we identify 8 molecular subgroups with distinct mutational and gene expression characteristics in adult MPAL. These results indicate that integrative genomic and transcriptomic profiling will provide informative implications on a more precise diagnosis, risk stratification and treatment options for MPAL.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal